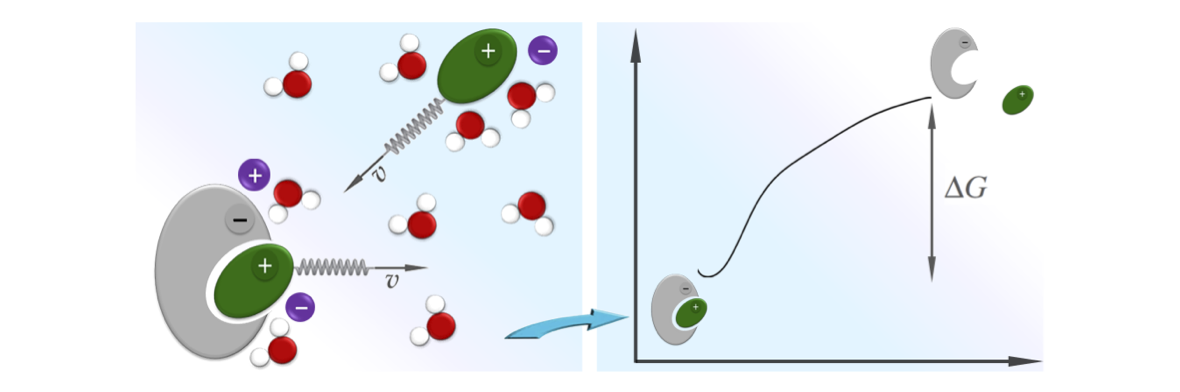
In collaboration with T. Do, P. Carloni, G. Varani, we have developed a novel method which can be used to accelerate atomistic molecular dynamics simulations of RNA/peptide binding. The trick is simple: since binding in this case is driven by electrostatic interactions, we use an estimate of the electrostatic interaction free-energy as an order parameter to monitor the binding event. The dynamics of this order parameter is then accelerated using e.g. steered molecular dynamics or other free-energy methods. This method has been applied to characterize the binding of TAR from HIV-1 and a small cyclic peptide of pharmaceutical relevance. Our study allowed to obtain a blind prediction of the binding site with a relatively low computational effort. As a side note, to better analyze pulling trajectories we have extended a recently proposed reweighting scheme. This allows to “pull” one order parameter and to “analyze” another one. We foresee the application of these novel techniques in the characterization of nucleic acids/peptide interaction.
This work is published as Do, Carloni, Varani, and Bussi, J. Chem. Theory Comput. 9, 1720 (2013), and is available free of charge. The preprint is available at arXiv:1307.5565
