Results
Our goals are twofold: (a) we develop new methodologies to improve molecular dynamics simulations and (b) we apply our techniques to relevant problems of RNA molecular biology. As such, also our results are twofold: Methods and Applications. A description of each of our publications can be found here. Alternatively, you can see all our papers indexed on Google Scholar.
Methods
We dedicate a lot of effort to the improvement of PLUMED, and we recently collaborated to the development of its latest version. PLUMED is a plugin that works in combination with several molecular dynamics engines providing features such as metadynamics and umbrella sampling with a wealth of different collective variables. Most of the methods we developed are thus included either in the official version of PLUMED or in separate, public branches.
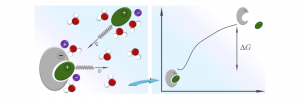
We have developed a method to accelerate binding simulations, that is particularly useful when studying charged molecules. Since RNA molecules are negatively charged and often bind positively charged proteins, we believe this method will be very helpful for the simulation of RNA/peptide binding.
We have also developed a very flexible version of the Hamiltonian replica exchange method, which can be used to accelerate selected portions of large molecules. The implementation is open source, integrated with PLUMED, and can be downloaded here.
We are currently working hard on other methods, results will be posted as soon as available.
Applications
One of our research lines is dedicated to the study of riboswitches. As a first step, we have characterized the ligand-induced stabilization in the add adenine riboswitch by means of accurate, atomistic molecular dynamics simulations. We also collaborated to a book chapter on the use of molecular docking on riboswitches.
We are currently working hard on other applications, results will be posted as soon as available.
